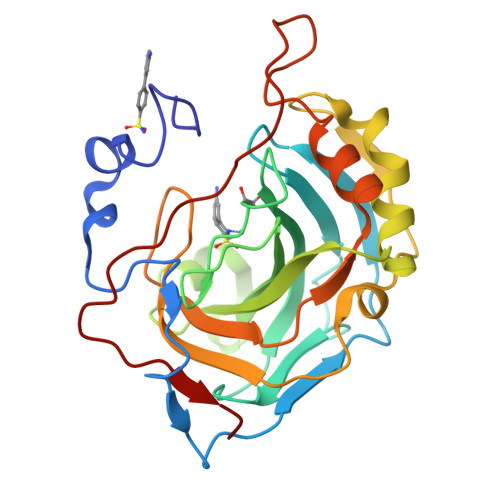
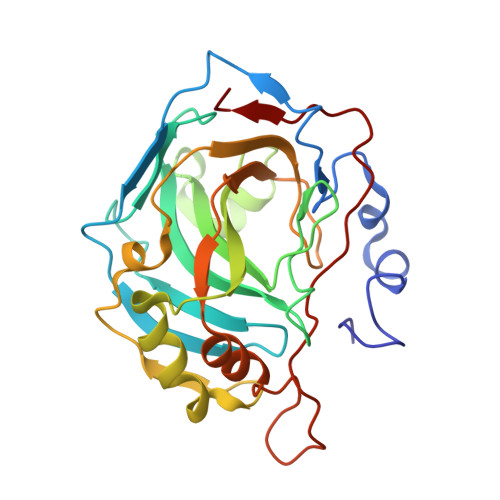
4-Arylbenzenesulfonamides as Human Carbonic Anhydrase Inhibitors (hCAIs): Synthesis by Pd Nanocatalyst-Mediated Suzuki-Miyaura Reaction, Enzyme Inhibition, and X-ray Crystallographic Studies.
Cornelio, B., Laronze-Cochard, M., Ceruso, M., Ferraroni, M., Rance, G.A., Carta, F., Khlobystov, A.N., Fontana, A., Supuran, C.T., Sapi, J.(2016) J Med Chem 59: 721-732
- PubMed: 26741028
- DOI: https://doi.org/10.1021/acs.jmedchem.5b01771
- Primary Citation of Related Structures:
5E28, 5E2K, 5E2S - PubMed Abstract:
Benzenesulfonamides bearing various substituted (hetero)aryl rings in the para-position were prepared by palladium nanoparticle-catalyzed Suzuki-Miyaura cross-coupling reactions and evaluated as human carbonic anhydrase (hCA, EC 4.2.1.1) inhibitors against isoforms hCA I, II, IX, and XII. Most of the prepared sulfonamides showed low inhibition against hCA I isoform, whereas the other cytosolic isoenzyme, hCA II, was strongly affected. The major part of these new derivatives acted as potent inhibitors of the tumor-associated isoform hCA XII. An opposite trend was observed for phenyl, naphthyl, and various heteroaryl substituted benzenesulfonamides which displayed subnanomolar hCA IX inhibition while poorly inhibiting the other tumor-associated isoform hCA XII. The inhibition potency and influence of the partially restricted aryl-aryl bond rotation on the activity/selectivity were rationalized by means of X-ray crystallography of the adducts of hCA II with several 4-arylbenzenesulfonamides.
Organizational Affiliation:
Institut de Chimie Moléculaire de Reims, CNRS UMR 7312, Université de Reims Champagne-Ardenne, UFR Pharmacie, 51 Rue Cognacq-Jay, F-51096 Reims Cedex, France.